|
snoRNA-LBME-db, a comprehensive database of
human H/ACA and C/D box snoRNAs.
VERSION 2
Introduction
C/D box snoRNPs
H/ACA box snoRNPs
Cajal body-specific scaRNAs
Telomerase RNA
Expression of snoRNAs
SnoRNAs and genomic imprinting
Selected reviews
Introduction
This database
of human C/D box and H/ACA modification guide RNAs is implemented at the Laboratoire
de Biologie Moléculaire Eucaryote (LBME, CNRS and Université Paul Sabatier)
in Toulouse, France, and maintained by Laurent Lestrade (Institut
d'Exploration Fonctionnelle des Génomes) et Michel Weber (Laboratoire
de Biologie Moléculaire Eucaryote).
We thank our colleagues at the LBME for their help in constructing this
database, and, in particular, Jean-Pierre Bachellerie for its compilation of snoRNA sequences.
Colleagues all around the world are encouraged to
address their criticisms, remarks and corrections to
Information on a particular snoRNA can be accessed
by three ways:
1-
On the Search page, just type the name of the snoRNA (for example ACA17)
in the Id window.
2-
The Find guide RNA contains the sequences of the human ribosomal rRNAs
28S, 18S and 5.8S, and of the snRNAs U1, U2, U4, U5 and U6, with the positions
of modified (2'O-ribose methylated or pseudo-uridinylated) nucleotides,
and the identity of the corresponding modification guide RNAs. You can
click on the name of the relevant snoRNA.
3-
This database is linked to the UCSC Genome
Browser. In the human genome Browser page, type the name of the snoRNA
(for example ACA17) in the "Position" window, and click on Submit.
You will be directed to the position of that snoRNA in the human genome.
Clicking on ACA17 on the right hand part of the Browser screen will direct
you to the corresponding page of the snoRNA-LBME-db.
The human rRNAs 28S, 18S and 5.8S carry together about
100 pseudouridines and 110 2'O-ribose methyl groups. These modifications
are catalysed by two families of ribonucleoparticules, the H/CA box and
C/D box snoRNPs, respectively. These two classes of snoRNPs are composed
of a commun set of four proteins, and a small RNA that specifically guides
the modification of one or two bases of the target RNA.
back to top
C/D box snoRNPs
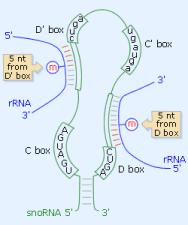
Figure from http://www.ergito.com/lookup.jsp?expt=kiss
|
The C/D box snoRNPs contain a commun set of four proteins: Fibrillarin
(FBL, the methyltransferase), NOP56 (or NOL5A, nucleolar protein 5A),
NOP5/NOP58 and NHPX (or NHP2L1, non-histone chromosome protein 2-like
1).
GeneLynx: FBL, NOL5A, NOP5/NOP58, NHP2L1
GeneCards:
FBL, NOL5A, NOP5/NOP58, NHP2L1
The C/D box snoRNAs carry the conserved boxes
C (RUGAUGA, R=purine) and D (CUGA) near their 5' and 3' ends, respectively.
The two boxes are frequently folded together by a short (4-5 bp) terminal
helix, to form a structure similar to a kink-turn. Often, imperfect
copies of the C and D boxes, named C' and D', are located internally,
in the order C/D'/C'/D. The 2'O-ribose methylation of target RNAs is
guided by one or two 10-21 anti-sens elements located upstream of the
D and/or D' boxes, so that the modified base is paired with the snoRNA
nucleotide located precisely 5 nts upstream of the D or D' box. |
back to top
H/ACA box snoRNPs
The H/ACA box small nucleolar (sno)RNPs contain a
commun set of four proteins: dyskerin (the pseudouridine synthase), GAR1
(nucleolar protein family A, member
1, NOLA1), NHP2 (NOLA2) and NOP10 (NOLA3).
GeneLynx: dyskerin, NOLA1, NOLA2, NOLA3
GeneCards: dyskerin, NOLA1, NOLA2,
NOLA3

Figure from http://www.ergito.com/lookup.jsp?expt=kiss |
The H/ACA box snoRNAs consist of two hairpins
and two short single-stranded regions, which contain the H box (ANANNA)
and the ACA box. The latter is always located 3 nts 5' of the 3' end
of the snoRNA (see Figure 1). The hairpins contain bulges, or recognition
loops, that form complex pseudo-knots with the target RNA, where the
target uridine is the first unpaired base (see Figure). The position
of the substrate uridine always resides 14-16 nts upstream of the H
box (left recognition pocket) or of the ACA box (right recognition
pocket). Some H/ACA box snoRNAs can thus guide the modification of
two uridines, sometimes in two different rRNAs (U69, ACA10, ACA31). However, many
H/ACA snoRNAs have only one identified target, and a growing number
have none.
|
back to top
Cajal body-specific scaRNAs
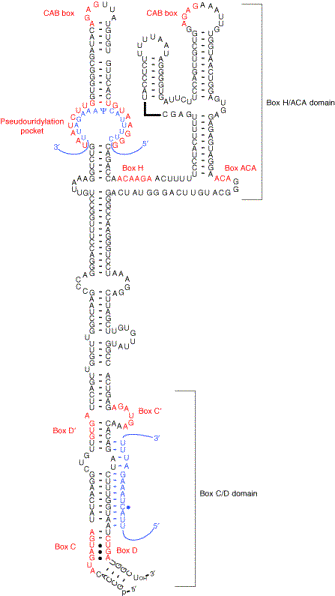 |
A new facet in H/ACA box small RNA's world
emerged with the cloning of U85, a 330 nt-long RNA
that contains both a C/D box and an H/ACA box domain. Accordingly,
U85 can guide both the pseudouridylation of base U46 and the 2'O-ribose
methylation of base C45 of the U5 snRNA. Subsequently, U85 was found
to co-localize with coilin in Cajal bodies.
U85 was the first member of the so-called small
Cajal body-specific (sca)RNAs. Other members have a C/D-H/ACA composite
structure similar to that of U85 (U87, U88, U89), while others have
only one (U92)
or two (U93)
H/ACA box domains. On the contrary, the scaRNAs U90 and U91 have the characteristic
structure of classical C/D box snoRNAs. The scaRNAs share three properties:
1- they co-localize with coilin in Cajal bodies.
2- they can guide modifications of the RNA polymerase
II-transcribed snRNAs (U1, U2, U4 and U5).
3- H/ACA box scaRNAs have one or two Cajal
body-specific localization signal, or CAB box (UGAG).
Reprinted from Henras, A.K., Dez, C. and Henry,
Y. Current Opinion in Structural Biology 14:335-343 (2004), with permission
from Elsevier. |
back to top
Telomerase RNA
The 3' most part of the human telomerase RNA (hTR or
TERC)) folds into a characteristic H/ACA box snoRNA-like structure. Accordingly,
hTR associates with the four core proteins of H/ACA box snoRNPs, dyskerin,
and NOLA1-3. Moreover, this H/ACA domain contains a Cajal body-specific
localization signal (CAB box), and localizes in Cajal bodies. Mutations
in hTR gene are the cause of the autosomal dominant form of dyskeratosis
congenita (OMIM#127550),
while mutations in the dyskerin (DCK1) gene cause the X-linked form of
the disease (OMIM#30500).
Expression of snoRNAs
In
vertebrates, sequences encoding H/ACA and C/D box snoRNAs and scaRNAs are
generally located in introns of their host gene, in the same orientation.
So far, an intron can carry only one snoRNA gene, but a host gene can carry
several snoRNA genes in different introns. However, the human C/D box snoRNAs
U3, U8, U13, mgU2-25/61 and mgU12-22/U4-8 are transcribed by
RNA polII as independent units. Intronic snoRNAs are produced by exonucleolytic
degradation of the debranched lariat after splicing, the stable part being
protected by the binding of snoRNP core proteins, and/or of ancillary proteins,
probably to the pre-mRNA.
The host genes of snoRNAs are either protein
coding or non-coding, and often belong to the family of 5' TOP (5'-Terminal
Oligo Pyrimidine tract) genes. Coding host genes include those of several
ribosomal proteins, or proteins associated with ribosome biosynthesis or
translation. Particularly interesting cases are the H/ACA box snoRNAs ACA36 and ACA56, that are located
in introns of the gene encoding dyskerin (one of the four core proteins
of H/ACA box snoRNPs). Similarly, the H/ACA box ACA51 and C/D box HBII-55 snoRNAs are
located in the gene encoding NOP56/NOL5A (one of the four core proteins
of C/D box snoRNPs). The C/D box snoRNA HBII-95 and HBII-234 are hosted
by the gene encoding NOP5/NOP58 protein (one of the four core proteins
of C/D box snoRNPs). However, many newly discovered snoRNAs reside in genes
with quite diverse functions, or of unknown function.
Intriguing cases are the six H/ACA box
snoRNAs ACA1, ACA8, ACA18, ACA25, ACA32 and ACA40, that reside
in the RefSeq gene MGC5306. This gene encodes an hypothetical protein of
278 aa. As snoRNAs in this gene are more conserved than exons, its main
function might be to produce snoRNAs rather than a protein. Similarly,
the C/D box snoRNAs U22, U25, U26, U27, U28, U29, U30 and U31 reside in introns
of the non-coding gene UHG (U22 host gene).
A growing number of snoRNAs have a tissue-specific
expression, most probably reflecting that of the host gene. This is the
case, for example, with the brain-specific H/ACA box snoRNA HBI-36,
that is located in the gene encoding the serotonin receptor 5HT-2c.
back to top
SnoRNAs and genomic imprinting
In the human and mouse genomes, two imprinted
loci contain clusters of tandemly arranged snoRNAs. At the human 15q11-q13
locus (Prader-Willi/Angelman syndrome region, OMIM#176270, OMIM#105830),
a very large (several hundreds of kb), maternally expressed and brain-specific
gene contains 2 tandems of 42 and 29 highly related copies of C/D snoRNAs, HBII-52 and HBII-85.
The human 14q32locus also contains a cluster of tandemly repeated, maternally
expressed, C/D box snoRNAs. The role, if any, of these snoRNAs in genomic
imprinting remains to be elucidated. However, the analysis of chromosomal
translocations has reduced the minimal interval for the Prader-Willi/Angelman
susceptibility region to a 121kb interval, in which the only known genes
are those encoding the cluster of C/D box snoRNAs HBII-85 and the unique
C/D box snoRNA HBII-438A (Gallagher et al.(2002)
Am. J. Hum. Genet. 71:669-678).
back to top
Selected reviews:
Pertinent original articles are given for
each snoRNA in the data base. Here are some selected reviews:
- Bachellerie, J. P., Cavaille, J., and Huttenhofer, A. (2002). The expanding snoRNA world. Biochimie, 84, 775-790
- Filipowicz, W., and Pogacic, V. (2002). Biogenesis of small nucleolar ribonucleoproteins. Curr Opin Cell Biol, 14, 319-327.
- Kiss, T. (2001). Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. Embo J, 20, 3617-3622.
- Kiss, T. (2002). Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell, 109, 145-148.
- Weinstein, L. B., and Steitz, J. A. (1999). Guided tours: from precursor snoRNA to functional snoRNP. Curr Opin Cell Biol, 11, 378-384.
- Henras, A. K., Dez, C., and Henry, Y. (2004). RNA structure and function in C/D and H/ACA s(no)RNPs. Curr Opin Struct Biol, 14, 335-343.
|


